
the recommended enrichmentmap version for Cytoscape 3.2 is the enrichmap version that contains the automatic annotation option (Release 2.1.0 ).more on java folders and set JAVA_HOME environment variable to Java 7 path.
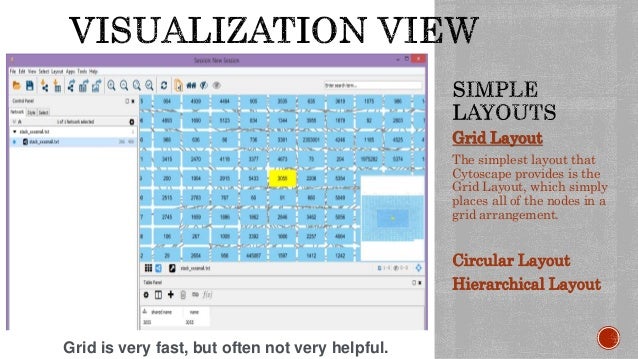
TOPOLOGICAL FEATURES USING CYTOSCAPE 3.2.1 HOW TO
Right click on the selected nodes and choose the option "group" => "group selected nodes" => name it.double click on the group on the node to collapse it.to expand a metanode, double-click on it and the individual nodes forming the group will be displayed.Īdd automatic labels using the "Annotate Cluster" feature (in menu -> EnrichmentMap -> Annotate Cluster).Run the "Annotate Cluster" with the option "Create Groups for clusters" checked (Control Panel -> Annotation Panel -> Advanced Option -> Create Groups for clusters).once the metanodes are created, we can change their size, color, shapes as other nodes.How to create a signature gene-set to use with the post-analysis option of enrichmentmap We also can import attributes for these nodes.take the list of genes that you want to overlap with the map.


 0 kommentar(er)
0 kommentar(er)
